PNGase F
Product information| Code | Name | Size | Quantity | Price | |
|---|---|---|---|---|---|
P0704S |
PNGase F |
15.000 units ( 500000 units/ml ) | - | Unavailable in your region | |
P0704L |
PNGase F |
75.000 units ( 500000 units/ml ) | - | Unavailable in your region |
PNGase F
Product Introduction
PNGase F is the most effective enzymatic method for removing almost all N-linked oligosaccharides from glycoproteins. PNGase F is an amidase, which cleaves between the innermost GlcNAc and asparagine residues of high mannose, hybrid, and complex oligosaccharides.
- Leaves N-glycan core oligosaccharides intact and suitable for further analysis
- Non-recombinant with no detectable endoglycosidase F1, F2 or F3 contamination
- ≥ 95% purity, as determined by SDS-PAGE and intact ESI-MS
- Stored in 50% glycerol
- Optimal activity and stability for up to 24 months
- Can be used under native or denaturing conditions
| Catalog # | Size | Concentration |
|---|---|---|
| P0704S | 15000 units | 500000 units/ml |
| P0704L | 75000 units | 500000 units/ml |
Featured Videos
View Video Library- Product Information
- Protocols, Manuals & Usage
- Tools & Resources
- FAQs & Troubleshooting
- Citations & Technical Literature
- Quality, Safety & Legal
- Other Products You May Be Interested In
Product Information
Description
Peptide -N-Glycosidase F, also known as PNGase F, is an amidase that cleaves between the innermost GlcNAc and asparagine residues of high mannose, hybrid, and complex oligosaccharides from N-linked glycoproteins (1)
Product Source
PNGase F is purified from Flavobacterium meningosepticum (3) and it is free of proteases and Endo F activities.Glycosidase Recognition Site
- This product is related to the following categories:
- Endoglycosidases,
- Proteome Analysis,
- This product can be used in the following applications:
- Expression Systems,
- Glycan Sequencing,
- Proteomics,
- Recombinant Glycoprotein Expression, Glycoprotein Analysis
Reagents Supplied
Reagents Supplied
The following reagents are supplied with this product:
| NEB # | Component Name | Component # | Stored at (°C) | Amount | Concentration | |
|---|---|---|---|---|---|---|
Properties & Usage
Unit Definition
One unit is defined as the amount of enzyme required to remove > 95% of the carbohydrate from 10 µg of denatured RNase B in 1 hour at 37°C in a total reaction volume of 10 µl.Unit Definition Assay:
10 µg of RNase B are denatured with 1X Glycoprotein Denaturing Buffer (0.5% SDS, 40 mM DTT) at 100°C for 10 minutes. After the addition of NP-40 and GlycoBuffer 2, two-fold dilutions of PNGase F are added and the reaction mix is incubated for 1 hour at 37°C. Separation of reaction products are visualized by SDS-PAGE.
1X Glycoprotein Denaturing Buffer
0.5% SDS
40 mM DTT
1X NP-40
1% NP-40 in MilliQ-H2O
Reaction Conditions
1X GlycoBuffer 2
Incubate at 37°C
1X GlycoBuffer 2
50 mM Sodium Phosphate
(pH 7.5 @ 25°C)
Storage Buffer
20 mM Tris-HCl
50 mM NaCl
5 mM EDTA
50% Glycerol
pH 7.5 @ 25°C
Heat Inactivation
75°C for 10 minutesMolecular Weight
Apparent: 36000 daltonsApplication Features
- Removal of high mannose N-glycans from glycoproteins
Related Products
Companion Products
Product Notes
- Since PNGase F activity is inhibited by SDS, it is essential to have NP-40 present in the reaction mixture. Why this non-ionic detergent counteracts the SDS inhibition is unknown at present.
- To deglycosylate a native glycoprotein, longer incubation time as well as more enzyme may be required.
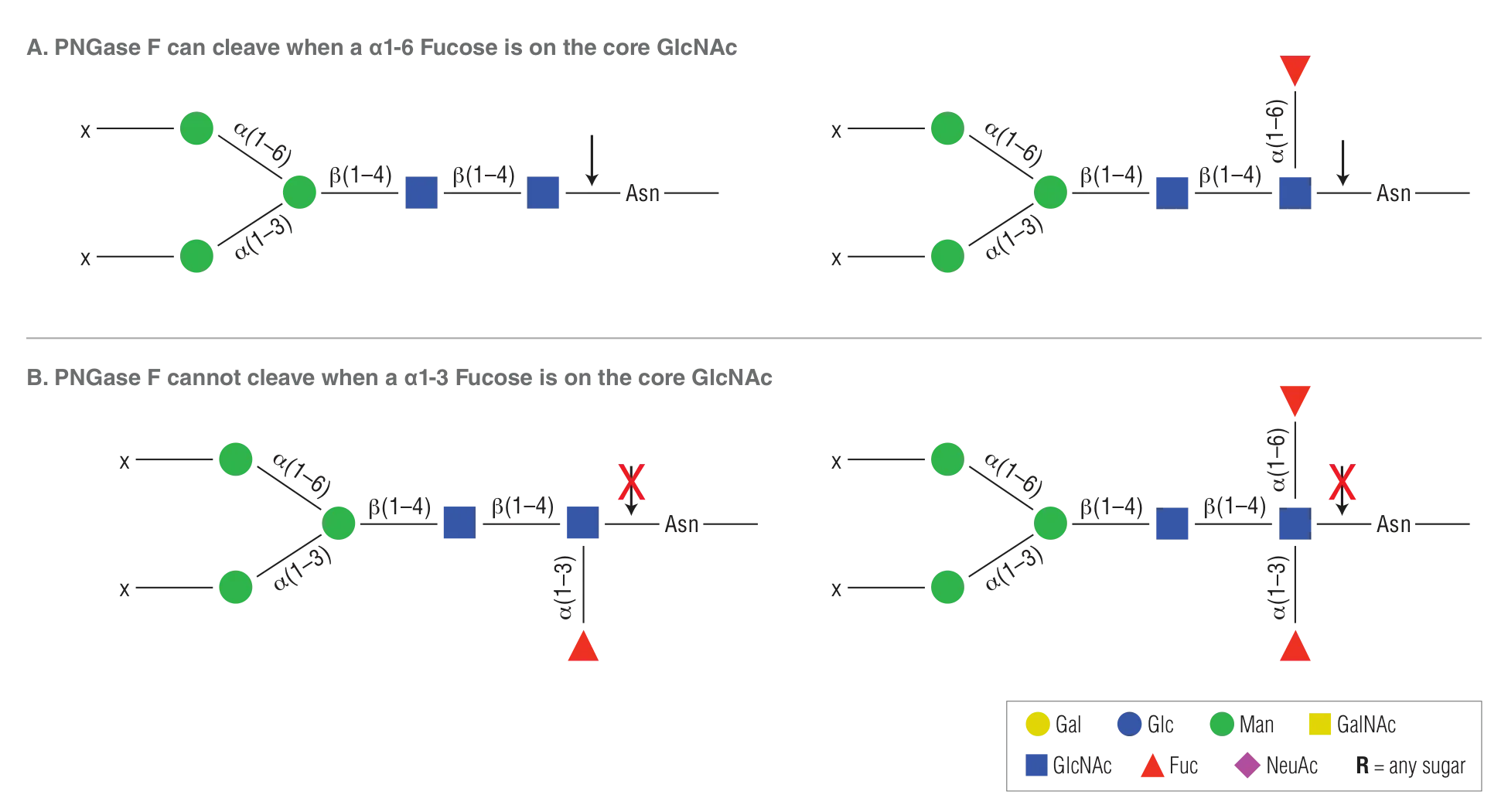
- PNGase F will not cleave N-linked glycans containing core α1-3 Fucose.
- Typical reaction conditions: Please see Protocols
References
- Maley, F. et al. (1989). Anal. Biochem. 180, 195-204.
- Tretter, V. et al. (1991). Eur. J. Biochem.. 199, 647-652.
- Plummer, T.H. Jr. and Tarentino, A.L. (1991). Glycobiology. 1, 257-263.
Protocols, Manuals & Usage
Protocols
Usage & Guidelines
Application Notes
- AppNote_Remove-iT_PNGase_F_Effective_Release_and_Recovery_of_Neutral_and_Sialylated_N-glycans
- AppNote_Glycan_Analysis_of_Murine_IgG_by_Enzymatic_Digestion_with_Endo_S_and_PNGase_F
- AppNote_Proteomics_Fast_and_Efficient_Antibody_Deglycosylation_using_Rapid_PNGase_F
- AppNote_Glycan_Analysis_of_Murine_IgG2a_by_Enzymatic_Digestion_with_PNGase_F_and_Trypsin
- Characterization of Glycans from Erbitux®, Rituxan® and Enbrel® using PNGase F (Glycerol-free), Recombinant
- Enzymatic removal of N- and O-glycans using PNGase F or the Protein Deglycosylation Mix
- Glycan Analysis of Murine IgG by Enzymatic Digestion with Endo S and PNGase F Followed by Mass Spectrometric Analysis
- Remove iT PNGase F Effective Release and Recovery of Neutral and Sialylated N glycans
- A Fast One-Step Digestion of DNA or RNA for Global Detection and Characterization of Nucleotide Modifications
- Glycan Analysis of Murine IgG2a by Enzymatic Digestion with PNGase F and Trypsin, Followed by Mass Spectrometric Analysis
Tools & Resources
Selection Charts
FAQs & Troubleshooting
FAQs
- Is PNGase F compatible with downstream analysis such as HPLC and Mass Spectrometry?
- What happens to the asparagine after PNGase removes the sugar?
- Why is my immunoprecipitated (IP) protein degraded? When I denature and add SDS all I see on my SDS-PAGE is a smear or no protein?
- What are the typical reaction conditions for PNGase F?
- Does PNGase F work in Urea?
- How do I inhibit PNGase F?
- How much PNGase F should I use to remove my carbohydrate under native conditions?
- I tried the PNGase F on my glycoprotein and didn't see removal of the carbohydrate. What could be the problem?
- What is the difference between PNGase F, Endo H and O-Glycosidase?
- Do detergents inhibit exoglycosidases/endoglycosidases?
- Why have the NEB Glycosidase enzymes changed reaction buffers? What are the new reaction buffers and can I still use an enzyme with its old buffer? Where can I find the composition of the old buffers?
- What are Glycosidases and their uses?
- What is a good endoglycosidase substrate?
Tech Tips
You can use this enzyme under native or denaturing conditions
Under native conditions we recommend adding more enzyme and using longer incubation times
PNGase F activity is inhibited by SDS, therefore under denaturing conditions it is essential to have NP-40 present in the reaction mixture in a 1:1 ratio.
PNGase F will not cleave N-linked glycans containing core a1-3 Fucose (PNGase A must be used in this instance)
Enzyme activity varies at different temperatures: 37°C - 100%; 30°C - 100%; 23°C - 65%; 17°C - 40% and 3°C - 0%
A good positive control substrate is RNase B
Citations & Technical Literature
Citations
Additional Citations
- Liu H., Zou X., Li T., Wang X., Yuan W., Chen Y., Han W. (2015) Enhanced production of secretory glycoprotein VSTM1-v2 with mouse IgGκ signal peptide in optimized HEK293F transient transfection Sci Rep; PubMedID: 26140918, DOI: 10.1038/srep11603
- Yang W., Zhang Y., Zhou X., Zhang W., Xu X., Chen R., Meng Q., Yuan J., Yang P., Yao B. (2015) Production of a Highly Protease-Resistant Fungal α-Galactosidase in Transgenic Maize Seeds for Simplified Feed Processing Sci Rep; 5, 11603. PubMedID: 26053048, DOI: 10.1038/srep11603
- Chen J., Fang M., Zhao YP., Yi CH., Ji J., Cheng C., Wang MM., Gu X., Sun QS., Chen XL., Gao CF. (2015) Serum N-Glycans: A New Diagnostic Biomarker for Light Chain Multiple Myeloma Sci Rep; 5, 11603. PubMedID: 26075387, DOI: 10.1038/srep11603
- Netsirisawan P., Chokchaichamnankit D., Srisomsap C., Svasti J., Champattanachai V. (2015) Proteomic Analysis Reveals Aberrant O-GlcNAcylation of Extracellular Proteins from Breast Cancer Cell Secretion Mol Biol Cell; 26, 2168-80. PubMedID: 26136220
- Brown EP., Normandin E., Osei-Owusu NY., Mahan AE., Chan YN., Lai JI., Vaccari M., Rao M., Franchini G., Alter G., Ackerman ME. (2015) Microscale purification of antigen-specific antibodies Sci Rep; PubMedID: 26078040, DOI: 10.1038/srep11603
- Shakiba N., White C.A., Lipsitz Y.Y., Yachie-Kinoshita A., Tonge P.D., Hussein S.M.I., Puri M.C., Elbaz J., Morrissey-Scoot J., Li M., Munoz J., Benevento M., Rogers I.M., Hanna J.H., Heck A.J.R., Wollscheid B., Nagy A., Zandstra P.W. (2015) CD24 tracks divergent pluripotent states in mouse and human cells. Nat Commun; 6, 7329. PubMedID: 26076835
- Malsburg K., Shao S., Hegde RS. (2015) The ribosome quality control pathway can access nascent polypeptides stalled at the Sec61 translocon Mol Biol Cell; 26, 2168-80. PubMedID: 25877867, DOI: 10.1091/mbc.E15-01-0040
- Olsen AL., Lai Y., Dalmau J., Scherer SS., Lancaster E. (2015) Caspr2 autoantibodies target multiple epitopes. Neurol Neuroimmunol Neuroinflamm; 2, e127. PubMedID: 26185774, DOI: 10.1212/NXI.0000000000000127
- Orizio F., Damiati E., Giacopuzzi E., Benaglia G., Pianta S., Schauer R., Schwartz-Albiez R., Borsani G., Bresciani R., Monti E. (2015) Human sialic acid acetyl esterase: Towards a better understanding of a puzzling enzyme. Glycobiology; 25, 992-1006. PubMedID: 26022516
- Singh S., Kuntal P., Yadav J., Tang H., Partyka K., Kletter D., Hsueh P., Ensink E., Kc B., Hostetter G., Xu E.H., Bern M., Smith D.F., Mehta A.S., Brand R., Melcher K., Haab B.B. (2015) Upregulation of glycans containing 3' fucose in a subset of pancreatic cancers uncovered using fusion-tagged lectins. J Proteome Res; 14, 2594-605. PubMedID: 25938165
- Go E.P., Herschhorn A., Gu C., Castillo-Menendez L., Zhang S., Mao Y., Chen H., Ding H., Wakefield J.K., Hua D., Liao H.X., Kappes J.C., Sodroski J., Desaire H. (2015) Comparative Analysis of the Glycosylation Profiles of Membrane-Anchored HIV-1 Envelope Glycoprotein Trimers and Soluble gp140. J Virol; 89, 8245-57. PubMedID: 26018173
- Haramoto Y., Takahashi S., Oshima T., Onuma Y., Ito Y., Asashima M. (2015) Insulin-like factor regulates neural induction through an IGF1 receptor-independent mechanism. Mol Biol Cell; 26, 2168-80. PubMedID: 26112133, DOI: 10.1091/mbc.E15-01-0040
- Wang L., Zhang X., Pang N., Xiao L., Li Y., Chen N., Ren M., Deng X., Wu J. (2015) Glycation of vitronectin inhibits VEGF-induced angiogenesis by uncoupling VEGF receptor-2-αvβ3 integrin cross-talk Mol Biol Cell; 26, 2168-80. PubMedID: 26111058, DOI: 10.1091/mbc.E15-01-0040
- Wang J., Hilchey SP., Hyrien O., Huertas N., Perry S., Ramanunninair M., Bucher D., Zand MS. (2015) Multi-Dimensional Measurement of Antibody-Mediated Heterosubtypic Immunity to Influenza. Sci Rep; 5, 11603. PubMedID: 26103163, DOI: 10.1038/srep11603
- Spelios MG., Olsen JA., Kenna LA., Akirav EM. (2015) Islet Endothelial Cells Induce Glycosylation and Increase Cell-surface Expression of Integrin β1 in β Cells Mol Biol Cell; 26, 2168-80. PubMedID: 25911095, DOI: 10.1091/mbc.E15-01-0040
- Pritchard L.K., Harvey D.J., Bonomelli C., Crispin M., Doores K.J. (2015) Cell- and Protein-Directed Glycosylation of Native Cleaved HIV-1 Envelope. J Virol; 89, 8932-44. PubMedID: 26085151
- Holemans T., Sørensen DM., Veen S., Martin S., Hermans D., Kemmer GC., Haute C., Baekelandt V., Pomorski TG., Agostinis P., Wuytack F., Palmgren M., Eggermont J., Vangheluwe P. (2015) A lipid switch unlocks Parkinson's disease-associated ATP13A2 Proc Natl Acad Sci U S A; 112, 9040-5. PubMedID: 26134396
- Asazuma HM., Sohn BH., Kim Y.S., Kuo CW., Khoo KH., Kucharski CA., Fraser MJ., Jarvis DL. (2015) Targeted Glycoengineering Extends the Protein N-glycosylation Pathway in the Silkworm Silk Gland Sci Rep; PubMedID: 26163436, DOI: 10.1038/srep11603
- AlSalmi W., Mahalingam M., Ananthaswamy N., Hamlin C., Flores D., Gao G., Rao V.B. (2015) A New Approach to Produce HIV-1 Envelope Trimers: BOTH CLEAVAGE AND PROPER GLYCOSYLATION ARE ESSENTIAL TO GENERATE AUTHENTIC TRIMERS. J Biol Chem; 290, 19780-95. PubMedID: 26088135
- Julien M., Chauvet S., Scheckenbach KE., Alfaidy N., Chanson M., Benharouga M. (2015) Involvement of the heterodimeric interface region of the nucleotide binding domain-2 (NBD2) in the CFTR quaternary structure and membrane stability Sci Rep; PubMedID: 26083625, DOI: 10.1038/srep11603
- Beata O., Jarząb A., Kratz E., Zimmer M., Gamian A., Ferens-Sieczkowska M. (2015) Terminal Mannose Residues in Seminal Plasma Glycoproteins of Infertile Men Compared to Fertile Donors Int J Mol Sci; 16, 14933-50. PubMedID: 26147424, DOI: 10.3390/ijms160714933
- Noble GP., Wang DW., Walsh DJ., Barone JR., Miller MB., Nishina KA., Li S., Supattapone S. (2015) A Structural and Functional Comparison Between Infectious and Non-Infectious Autocatalytic Recombinant PrP Conformers Sci Rep; 5, 11603. PubMedID: 26125623, DOI: 10.1038/srep11603
- Itahana Y, Han R, Barbier S, Lei Z, Rozen S, Itahana K (2014) The uric acid transporter SLC2A9 is a direct target gene of the tumor suppressor p53 contributing to antioxidant defense Oncogene; PubMedID: 24858040, DOI: 10.1038/onc.2014.119
- Rosenbaek LL, Kortenoeven ML, Aroankins TS, Fenton RA (2014) Phosphorylation decreases ubiquitylation of the thiazide-sensitive cotransporter NCC and subsequent clathrin-mediated endocytosis J Biol Chem; 289(19), 13347-61. PubMedID: 24668812, DOI: 10.1074/jbc.M113.543710
- Botto L, Cunati D, Coco S, Sesana S, Bulbarelli A, Biasini E, Colombo L, Negro A, Chiesa R, Masserini M, Palestini P (2014) Role of lipid rafts and GM1 in the segregation and processing of prion protein PLoS One; 9(5), e98344. PubMedID: 24859148, DOI: 10.1371/journal.pone.0098344
- Wright CR, Brown EL, Della-Gatta PA, Ward AC, Lynch GS, Russell AP (2014) G-CSF does not influence C2C12 myogenesis despite receptor expression in healthy and dystrophic skeletal muscle Front Physiol; 5, 170. PubMedID: 24822049, DOI: 10.3389/fphys.2014.00170
- Wicht O, Burkard C, de Haan CA, van Kuppeveld FJ, Rottier PJ, Bosch BJ (2014) Identification and Characterization of a Proteolytically Primed Form of the Murine Coronavirus Spike Proteins after Fusion with the Target Cell J Virol; 88(9), 4943-52. PubMedID: 24554652, DOI: 10.1128/JVI.03451-13
- Kwon HM, Lee KH, Han BW, Han MR, Kim DH, Kim DE (2014) An RNA aptamer that specifically binds to the glycosylated hemagglutinin of avian influenza virus and suppresses viral infection in cells PLoS One; 9(5), e97574. PubMedID: 24835440, DOI: 10.1371/journal.pone.0097574
- Stech M, Quast RB, Sachse R, Schulze C, Wüstenhagen DA, Kubick S (2014) A continuous-exchange cell-free protein synthesis system based on extracts from cultured insect cells PLoS One; 9(5), e96635. PubMedID: 24804975, DOI: 10.1371/journal.pone.0096635
- Haller G, Li P, Esch C, Hsu S, Goate AM, Steinbach JH (2014) Functional characterization improves associations between rare non-synonymous variants in CHRNB4 and smoking behavior PLoS One; 9(5), e96753. PubMedID: 24804708, DOI: 10.1371/journal.pone.0096753
Quality, Safety & Legal
Quality Assurance Statement
Quality Control tests are performed on each new lot of NEB product to meet the specifications designated for it. Specifications and individual lot data from the tests that are performed for this particular product can be found and downloaded on the Product Specification Sheet, Certificate of Analysis, data card or product manual. Further information regarding NEB product quality can be found here.Specifications
The Specification sheet is a document that includes the storage temperature, shelf life and the specifications designated for the product. The following file naming structure is used to name these document files: [Product Number]_[Size]_[Version]Certificate Of Analysis
The Certificate of Analysis (COA) is a signed document that includes the storage temperature, expiration date and quality controls for an individual lot. The following file naming structure is used to name these document files: [Product Number]_[Size]_[Version]_[Lot Number]- P0704S_L_v1_0431603
- P0704S_L_v1_0431610
- P0704S_L_v1_0421504
- P0704S_L_v1_0421507
- P0704S_L_v1_0431703
- P0704S_L_v1_0431712
- P0704L_v1_10009228
- P0704S_v1_10015230
- P0704L_v1_10015218
- P0704L_v1_10021693
- P0704S_v1_10021692
- P0704S_v1_10027171
- P0704L_v1_10029401
- P0704L_v1_10032357
- P0704S_v1_10031035
- P0704L_v1_10034947
- P0704S_v1_10034949
- P0704S_v1_10039550
- P0704L_v1_10043786
- P0704S_v1_10047993
- P0704L_v1_10050897
- P0704S_v1_10050898
- P0704L_v1_10057010
- P0704S_v1_10057011
- P0704L_v1_10061011
- P0704S_v1_10061027
- P0704L_v1_10063053
- P0704L_v1_10067818
- P0704S_v1_10063054
- P0704L_v1_10071837
- P0704L_v1_10078698
- P0704S_v1_10071840
- P0704L_v1_10081144
- P0704S_v1_10081145
- P0704L_v1_10085486
- P0704S_v1_10085488
- P0704S_v1_10092595
- P0704S_v1_10094766
- P0704L_v1_10094767
- P0704S_v1_10098276
- P0704S_v1_10105452
- P0704L_v1_10105453
- P0704L_v1_10111500
- P0704S_v1_10111498
- P0704S_v1_10118379
- P0704L_v1_10118380
- P0704L_v1_10129278
- P0704S_v1_10129277
- P0704L_v1_10134387
- P0704S_v1_10139346
- P0704L_v1_10139347
- P0704S_v1_10141860
- P0704S_v1_10151808
- P0704L_v1_10151807
- P0704L_v1_10161533
- P0704L_v1_10164658
- P0704S_v1_10164657
- P0704S_v1_10171475
- P0704L_v1_10171476
- P0704S_v1_10178736
- P0704L_v1_10189740
- P0704S_v1_10190548
- P0704L_v1_10203687
- P0704S_v1_10203686
- P0704S_v1_10218796
- P0704L_v1_10214338
- P0704L_v1_10231431
- P0704S_v1_10231463
- P0704L_v1_10241656
- P0704S_v1_10244094
- P0704L_v1_10250774
- P0704L_v1_10262263
- P0704S_v1_10270379
- P0704L_v1_10269837
- P0704S_v1_10274119
- P0704L_v2_10284996
- P0704S_v2_10287385
- P0704L_v2_10289549
- P0704S_v2_10301760
- P0704L_v2_10303200
Safety DataSheets
The following is a list of Safety Data Sheet (SDS) that apply to this product to help you use it safely.PNGase F
GlycoBuffer 2
Glycoprotein Denaturing Buffer
NP-40
Legal and Disclaimers
Products and content are covered by one or more patents, trademarks and/or copyrights owned or controlled by New England Biolabs, Inc (NEB). The use of trademark symbols does not necessarily indicate that the name is trademarked in the country where it is being read; it indicates where the content was originally developed. The use of this product may require the buyer to obtain additional third-party intellectual property rights for certain applications. For more information, please email busdev@neb.com.This product is intended for research purposes only. This product is not intended to be used for therapeutic or diagnostic purposes in humans or animals.
New England Biolabs (NEB) is committed to practicing ethical science – we believe it is our job as researchers to ask the important questions that when answered will help preserve our quality of life and the world that we live in. However, this research should always be done in safe and ethical manner. Learn more.
Other Products You May Be Interested In
The supporting documents available for this product can be downloaded below.








