Endo H
Product information| Code | Name | Size | Quantity | Price | |
|---|---|---|---|---|---|
P0702S |
Endo H |
10.000 units ( 500000 units/ml ) | - | Unavailable in your region | |
P0702L |
Endo H |
50.000 units ( 500000 units/ml ) | - | Unavailable in your region |
Endo H
New Reaction Buffer
Product Introduction
Endoglycosidase H is a recombinant glycosidase which cleaves within the chitobiose core of high mannose and some hybrid oligosaccharides from N-linked glycoproteins. A tagged version, which is a recombinant protein fusion of Endoglycosidase H and maltose binding protein is also available, Endo Hf (NEB #P0703).
- Recombinant enzyme with no detectable exoglycosidase or other endoglycosidase contaminating activities
- Glycerol-free for optimal performance in HPLC and mass spectrometry analysis
- ≥95% purity, as determined by SDS-PAGE and intact ESI-MS
- Optimal activity and stability for up to 24 months
| Catalog # | Size | Concentration |
|---|---|---|
| P0702S | 10000 units | 500000 units/ml |
| P0702L | 50000 units | 500000 units/ml |
- Product Information
- Protocols, Manuals & Usage
- Tools & Resources
- FAQs & Troubleshooting
- Citations & Technical Literature
- Quality, Safety & Legal
- Other Products You May Be Interested In
Product Information
Description
Endoglycosidase H is a recombinant glycosidase which cleaves within the chitobiose core of high mannose and some hybrid oligosaccharides from N-linked glycoproteins.


Product Source
Cloned from Streptomyces plicatus (2) and overexpressed in E.coli (3).- This product is related to the following categories:
- Endoglycosidases,
- Proteome Analysis,
- This product can be used in the following applications:
- Expression Systems,
- Glycan Sequencing,
- Proteomics,
- Recombinant Glycoprotein Expression, Glycoprotein Analysis
Reagents Supplied
Reagents Supplied
The following reagents are supplied with this product:
| NEB # | Component Name | Component # | Stored at (°C) | Amount | Concentration | |
|---|---|---|---|---|---|---|
Properties & Usage
Unit Definition
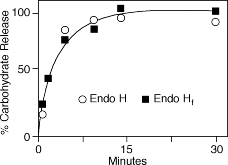
One unit is defined as the amount of enzyme required to remove > 95% of the carbohydrate from 10 µg of denatured RNase B in 1 hour at 37°C in a total reaction volume of 10 µl (10 NEB units = 1 IUB milliunit).Unit Definition Assay:
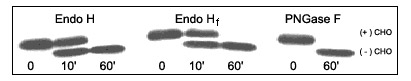
10 µg of RNase B are denatured with 1X Glycoprotein Denaturing Buffer at 100°C for 10 minutes. After the addition of 1X GlycoBuffer 3, two-fold dilutions of Endo H are added and the reaction mix is incubated for 1 hour at 37°C. Separation of reaction products is visualized by SDS-PAGE.
1X Glycoprotein Denaturing Buffer
0.5% SDS
40 mM DTT
Reaction Conditions
1X GlycoBuffer 3
Incubate at 37°C
1X GlycoBuffer 3
50 mM sodium acetate
(pH 6 @ 25°C)
Storage Buffer
20 mM Tris-HCl
50 mM NaCl
5 mM EDTA
pH 7.5 @ 25°C
Heat Inactivation
75°C for 10 minutesMolecular Weight
Apparent: 29 kDaApplication Features
- Removal of high mannose N-glycans from glycoproteins
Related Products
Companion Products
Product Notes
- Enzymatic activity is not affected by SDS.
- To deglycosylate a native glycoprotein, longer incubation time as well as more enzyme may be required.
- Activity at different temperatures (measured after a 1 hour incubation of glycosidase and denatured RNase B at the given temperature): 37°C - 100%; 30°C - 65%; 25°C - 40%; 17°C - 25% and 2°C - 0%.
- Typical reaction conditions: Please see FAQs.
References
- Maley, F. et al. (1989). Anal. Biochem. 180, 195-204.
- Robbins, P. et al. (1984). J. Biol. Chem. 259, 7577-7583.
- Guan, C. and Wong,S. New England Biolabs, unpublished observations.
Protocols, Manuals & Usage
Protocols
Tools & Resources
Selection Charts
FAQs & Troubleshooting
FAQs
- What is the difference between PNGase F, Endo H and O-Glycosidase?
- What is the difference between Endo H and Endo Hf?
- I tried the Endo H/Hf on my glycoprotein and it failed. What could be the problem?
- How much Endo H/Endo Hf should I use?
- Is EndoH/ Endo Hf inhibited by SDS?
- What are the typical reaction conditions for Endo H?
- Are Protease Inhibitors acceptable for use in an Endo H/Hf reaction?
- What are Glycosidases and their uses?
- Why have the NEB Glycosidase enzymes changed reaction buffers? What are the new reaction buffers and can I still use an enzyme with its old buffer? Where can I find the composition of the old buffers?
- What is a good endoglycosidase substrate?
- Do detergents inhibit exoglycosidases/endoglycosidases?
Tech Tips
Citations & Technical Literature
Citations
Additional Citations
- Arakel EC, Brandenburg S, Uchida K, Zhang H, Lin YW, Kohl T, Schrul B, Sulkin MS, Efimov IR, Nichols CG, Lehnart SE, Schwappach B (2014) Tuning the electrical properties of the heart by differential trafficking of KATP ion channel complexes J Cell Sci; 127(Pt 9), 2106-19. PubMedID: 24569881, DOI: 10.1242/jcs.141440
- Möykkynen T, Coleman SK, Semenov A, Keinänen K (2014) The N-terminal domain modulates α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) receptor desensitization J Biol Chem; 289(19), 13197-205. PubMedID: 24652293, DOI: 10.1074/jbc.M113.526301
- Rosenbaum EE, Vasiljevic E, Brehm KS, Colley NJ (2014) Mutations in four glycosyl hydrolases reveal a highly coordinated pathway for rhodopsin biosynthesis and N-glycan trimming in Drosophila melanogaster PLoS Genet; 10(5), e1004349. PubMedID: 24785692, DOI: 10.1371/journal.pgen.1004349
Quality, Safety & Legal
Quality Assurance Statement
Quality Control tests are performed on each new lot of NEB product to meet the specifications designated for it. Specifications and individual lot data from the tests that are performed for this particular product can be found and downloaded on the Product Specification Sheet, Certificate of Analysis, data card or product manual. Further information regarding NEB product quality can be found here.Specifications
The Specification sheet is a document that includes the storage temperature, shelf life and the specifications designated for the product. The following file naming structure is used to name these document files: [Product Number]_[Size]_[Version]Certificate Of Analysis
The Certificate of Analysis (COA) is a signed document that includes the storage temperature, expiration date and quality controls for an individual lot. The following file naming structure is used to name these document files: [Product Number]_[Size]_[Version]_[Lot Number]- P0702S_L_v1_0181603
- P0702S_L_v1_0181611
- P0702S_L_v1_0191612
- P0702S_L_v1_0191708
- P0702S_L_v1_0191802
- P0702L_v1_10021981
- P0702S_v1_10021982
- P0702L_v1_10040021
- P0702S_v1_10040019
- P0702L_v1_10048769
- P0702S_v1_10040023
- P0702S_v1_10048770
- P0702L_v1_10056020
- P0702S_v1_10062212
- P0702L_v1_10062668
- P0702L_v1_10069239
- P0702S_v1_10077476
- P0702S_v1_10081123
- P0702L_v1_10086145
- P0702S_v1_10086146
- P0702S_v1_10096798
- P0702L_v1_10096801
- P0702S_v1_10120777
- P0702L_v1_10124276
- P0702S_v1_10124277
- P0702S_v1_10151122
- P0702S_v1_10163719
- P0702L_v1_10163718
- P0702L_v1_10181109
- P0702S_v1_10181432
- P0702S_v1_10209944
- P0702L_v1_10209950
- P0702S_v1_10245971
- P0702L_v1_10246132
- P0702L_v1_10280133
- P0702S_v1_10281718
- P0702S_v1_10292874
- P0702S_v1_10307596
Safety DataSheets
The following is a list of Safety Data Sheet (SDS) that apply to this product to help you use it safely.Endo H
GlycoBuffer 3
Glycoprotein Denaturing Buffer
Legal and Disclaimers
Products and content are covered by one or more patents, trademarks and/or copyrights owned or controlled by New England Biolabs, Inc (NEB). The use of trademark symbols does not necessarily indicate that the name is trademarked in the country where it is being read; it indicates where the content was originally developed. The use of this product may require the buyer to obtain additional third-party intellectual property rights for certain applications. For more information, please email busdev@neb.com.This product is intended for research purposes only. This product is not intended to be used for therapeutic or diagnostic purposes in humans or animals.
New England Biolabs (NEB) is committed to practicing ethical science – we believe it is our job as researchers to ask the important questions that when answered will help preserve our quality of life and the world that we live in. However, this research should always be done in safe and ethical manner. Learn more.
Other Products You May Be Interested In
The supporting documents available for this product can be downloaded below.






